

- #Files edited with 4 peaks not opening sequencher for free
- #Files edited with 4 peaks not opening sequencher software download
- #Files edited with 4 peaks not opening sequencher software
Use this software to perform DNA fragment analysis, separate a mixture of DNA fragments according to their sizes, provide a profile of the separation, and precisely calculate the sizes of the fragments.
#Files edited with 4 peaks not opening sequencher for free
Peak Scanner Software is a DNA sizing software that can either be downloaded for free or purchased for free as a software kit.
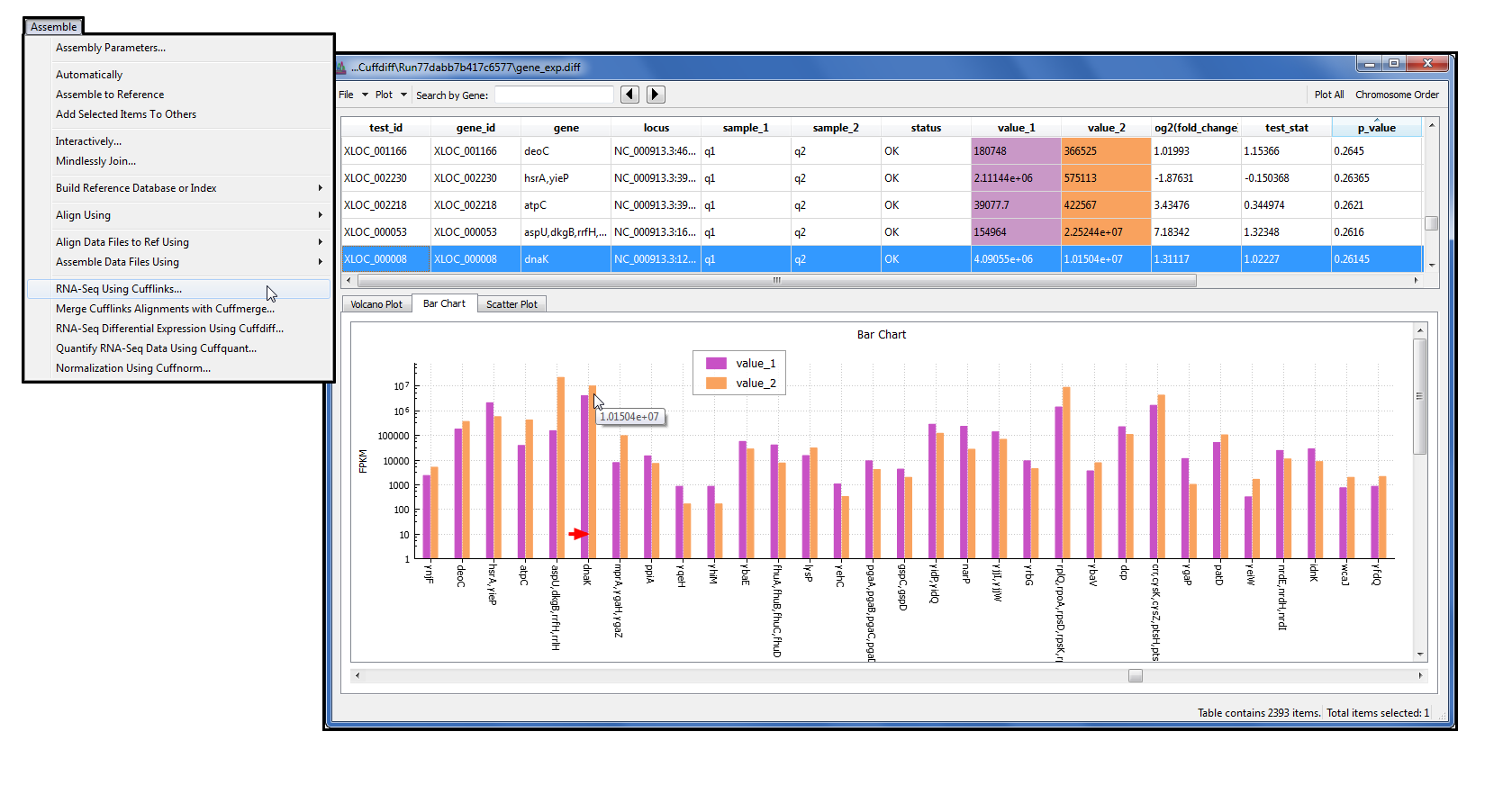
Microsatellite Analysis is commonly used for Microsatellite Instability in cancer, Triplet Repeat Expansion in neurodegenerative diseases, Species Identification & Characterization and Human Sample Authentication. This analysis provides a profile of the separation, precisely calculates the sizes of the fragments, and determines the microsatellite alleles present in the sample. Microsatellite Analysis Software(MSA) is a microsatellite genotyping software that allows you to analyze a mixture of DNA fragments, separated by size. The software allows you to view, edit, analyze, print, and export fragment analysis data generated using Applied Biosystems genetic analyzers. Peak Scanner module is a DNA fragment sizing software that performs DNA fragment analysis, separate a mixture of DNA fragments according to their sizes, provide a profile of the separation, and precisely calculate the sizes of the fragments. Sizing Analysis Module, Peak Scanner Software The Applied Biosystems SeqScreener Gene Edit Confirmation App (SGC) is a free and user-friendly software to determine the range and frequency of mutations generated in CRISPR-Cas9 experiments. SeqScreener Gene edit confirmation app (SGC) There is no need for local database setup, so computer resources are easily developed. MicrobeBridge Software is a streamlined, desktop software solution that connects DNA sequences generated on Applied Biosystems Sanger sequencers with the Centers for Disease Control and Prevention (CDC)’s MicrobeNet™ database for bacterial identification using 16S rRNA gene sequencing analysis. The software analyzes data generated using an Applied Biosystems MicroSEQ chemistry kit and an Applied Biosystems capillary-based genetic analyzer. MicroSEQ ID Microbial Identification Software is a tool for identification of bacteria and fungi. MicroSEQ ID Microbial Identification Software The results are automatically annotated with known SNPs from the current genomic database. The NGC module provides fast analysis of AB1 files and reports variants in genomic coordinates. The Next-Generation Confirmation (NGC) Module lets users compare results from standard NGS variant files with results from Sanger sequencing instruments within the Thermo Fisher Cloud environment.Ĭritical decisions often require validation of NGS results using robust Sanger sequencing. Next Generation Confirmation (NGC) Module There is no software maintenance required from users. The VA Module also reports and exports variant files in standard. With highly overlapped forward/reverse strands, the VA Module reports very high sensitivity for SNP calls. The VA Module can automatically retrieve reference sequences from the genomic database, report variants with genomic coordinates, and report genomic annotations for SNPs. The Variant Analysis (VA) Module provides fast analysis of Sanger sequencing data. The robust algorithms will call SNPs, mutations, insertions, deletions, and heterozygous insertions⁄deletions for data generated using Applied Biosystems genetic analyzers. Variant Reporter Software is designed for reference-based and non-reference-based analysis such as mutation detection and analysis, SNP discovery and validation, and sequence confirmation.
#Files edited with 4 peaks not opening sequencher software download
Variant Reporter Software Download trial version › It provides library functions for comparison to a known group of sequences, as well as features to assist with 21 CFR Part 11 compliance (Security, Audit and electronic signature features), which can be important in clinical research labs. SeqScape Software is a resequencing package designed for mutation detection and analysis, SNP discovery and validation, pathogen sub-typing, allele identification, and sequence confirmation. Windows™ 7 SP1, 32-bit or 64-bit or Windows™ 10 The improved sensitivity makes Sanger sequencing a fast, cost effective, and accurate way to call low-frequency somatic variants where the number of relevant targets is limited.
Minor Variant Finder Software enables 5% somatic variant detection using Sanger sequencing.


 0 kommentar(er)
0 kommentar(er)
